DxSeq

Interpretation and analysis of the NGS data
DxSeq provides NGS data analysis for a variety of genomic applications including targeted resequencing, exome and whole genome sequencing.
With robust tools for cancer gene panels and human disease diagnostics, you can complete your entire analysis on a single platform.
| Input | Fast Files |
|---|---|
| Output |
QC File, BAM File Excel Annotation/Interpretation file with multiple sheets(pathogenic variants, all variants, Gene CNV, info) Visualized Chromosomal CNV files(whole genome view, single chromosome view) Visualized Gene CNV files PDF Report Pages(if requested)* |
Workflow
Alignment
SNP/Indels Callings
Annotation
Filtering
ACMG Classification
DxSeq Benefits
- Cloud-based
- Accessible online, anywhere, anytime
- User-friendly interface
- Variant pathogenicity interpretation based on ACMG guideline
- Analysis of Paired/Single End data
- Batch management and analysis of multiple samples in the batch context
- Advanced options for customized filtering/annotation
- QC file development
- Comprehensive annotation (population/diseases databases and predictive tools)
- SNP Detection
- Indel Detection
- Gene/Exon Copy Number Variation (CNV) detection
- Chromosomal CNV detection
- Low False Positive Rate
- Automatic final report page generation
- Applicable to any Targeted Gene Panel
Key Features

Comprehensive Annotation
Helps you see as much information as possible in a glance, including ACMG criteria and population/diseases databases and predictive tools.
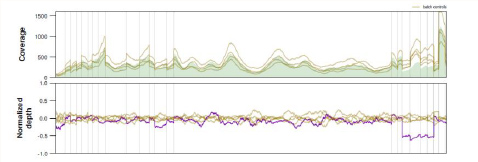
Gene CNVs
Visualized gene CNV analysis for each and every Gene in the Panel illustrating Gene/Exon Copy Number Variations
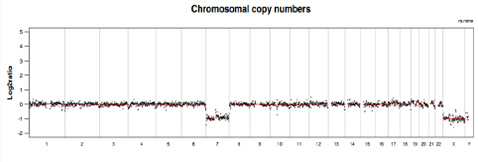
Chromosomal CNVs
Visualized chromosomal CNV analysis illustrated in genome plot and 23 single chromosome plots
